Detecting bacterial Pathogens and Antibiotic Resistances with Split Ribozymes
©spear © igem hamburg
Antibiotic resistant germs are an ever-increasing problem of which all of us have already heard. There are different approaches to solve the problem:
- informing patients and doctors, so that less antibiotics are prescribed
- environment and animal protection laws to reduce the prophylactic use of antibiotics
- research on new antibiotic against which there no resistances yet
- research on better and faster tests
All strategies combined will hopefully eventually lead to the achievement of the goal. However, without a fast and accurate evidence of the pathogens, we will not be able to solve the problem.
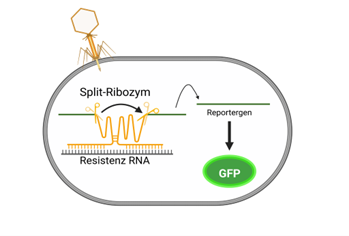
And that is when we enter the game: SPEAR - Sensing Pathogens and Emerging Antibiotic Resistances. We are a student research team of bachelor- und master students studying Molecular Life Sciences and Nano Sciences. Our goal is to prove the existence of bacteria and their resistances. There are many possibilities to do it. However, most of them require a lot of lab equipment and which are very specific or just take a lot of time because they are based on the growth of bacteria. We intend to develop a faster and easier test and thus focus on the RNA of the bacteria as well as the RNA, which they need to develop a resistance. We are using a split-ribozyme. Ribozyme are RNAs, which can cut themselves out (autocatalytic). If we place them in front of a reporter gene, we can see the colored reporter only if the ribozyme has cut itself out. In our case the ribozyme is divided and inactive in this state, i. e., we do not see a reporter. It will only be activated if it connects with another specific RNA and can cut itself out and we can see the reporter. This system is rather simple to adapt to all possible RNAs in order to be able to prove a wide spectrum of bacteria and their corresponding resistances. We insert our split-ribozyme into the cells by using phages because they are easy to store (thanks to the vaccinations, we all know that the storage of RNA is rather complicated). In our project, we work with harmless bacteria exclusively, the well-researched Escherichia coli in order to make a proof-of-concept for our idea.
Further information and details can be found at our Wiki:
https://2022.igem.wiki/uni-hamburg/index.html
©spear
figure: Resistant bacteria produce the fluorescent reporter GFP (Created with BioRender.com)
Student research group
- Stine Marla Behrmann
- Jonas Westphal
- Jeoline Rehnert
Mentor
- Prof. Michael Kolbe


